Repetitive scaffolds
Proteins with specific molecular recognition are invalauable tools used in a wide range of biotechnological and biomedical applications. The most common binding molecules are high affinity and specificity proteins – antibodies. However , due to its limitations: large size, disulphide bonds, low stability; there is a great need for alternative nonimmunoglobulin scaffolds. So far many different structural scaffolds have been employed as an antibody replacement – starting from Ig-like fold molecules (anticalins) through repetitive proteins (DARPins) to small helical bundle (Affibody). They have already been proven to be useful binders for different molecular targets. We have proposed three novel alternative scaffolds for generating affinity binders by Phage Display technology:
M3α
Small, (50 aminoacid) stable, helical domain – named by us a M3α ,with high structural similarity to Affibody scaffold has been proposed as an alternative binder structure.
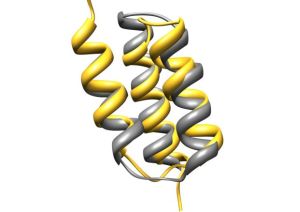
Structural similarity of Affibody and M3α. Grey color represent the structure of M3α, PDB 1oks; yellow corresponds to structure of Affibody PDB 1lp1 chain B. Superposition was done by Chimera software (Q score 0,62).

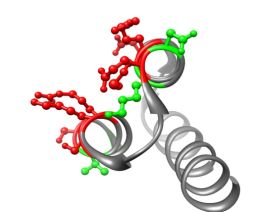
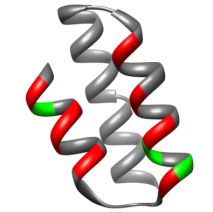
On the basis of structures of Affibody with its ligands, protein-protein interaction aminoacid positions have been chosen. Different types of randomization have been proposed: hard randomization NNC – in 8 position in the binding core; partially randomization – KMT and HWC – in 11 postions. It was decided to use both conventional – Sec and SRP translocation pathways.
Structure of M3α with randomized position highlated. Positions in red correspond to all libraries, those in green only to partially randomised: C4, S4, C6, S6 libraries.
| Library | Translocation pathway | Randomization | Theoretical size |
| C4 | Sec – pelB | KMT – Y/S/A/D 11 positions | 4 11 4 x 106 |
| S4 | SRP – DsbA | ||
| C6 | Sec – pelB | HWC – N/I/H/L/Y/F 11 positions | 6 11 8 x 109 |
| S6 | SRP – DsbA | ||
| C15 | Sec – pelB | NNC – A/C/D/F/G/H/I/L/N/P/R/S/T/V/Y 8 positions | 15 8 2 x 109 |
| S15 | SRP – DsbA |
TPR
TPR (tetratricopeptide repeat) is a structural motif mediating protein-protein interactions and assembly of multi-protein complexes in a number of different organisms. The basic repeat is comprised of 34 amino acids and forms a helix – turn – helix structure. TPR domains in proteins consist of 3-16 TPR motifs (adjacent TPRs packed in a parallel fashion), however 3 tandemly arrayed TPR motifs are the most prevalent in proteins and represent probably the minimal number of repeats required for specific ligand binding.
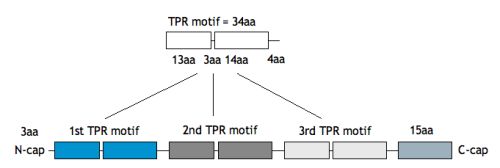
We have chosen the 3-TPR motif as potential protein scaffold. Based mainly on individual TPR sequences available at SMART and Pfam databases, sequence alignment has been done, revealing evolutionary conserved positions in consensus sequence. Some rational design has been applied to neutralized the overall charge of the design TPR (DTPR). Additional N- and C-capping sequences have been added to increased the stability and solubility of the construct (total length of 120aa).
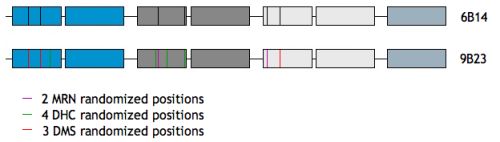
The phage display (PD) libraries were designed based on analysis and comparison of crystal structures of TPR complexes with ligands (PDB 1elr, 1elw, 2c2l, 1fch, 2qfc, 2uwj, 1na0). Protein-protein interaction amino-acid residues at internal concave surface have been chosen and 2 types of libraries have been proposed:
6B14 – 6 residues, each randomized to 14 amino-acids, resulting in size of 7.5×106
9B23 – 9 residues, DMS, DHC, MRN types of randomization, amino-acid size of 1.2×108
As the scaffold is extremely stable and helical we applied the co-translational SRP (signal recognition particle) protein translocation pathway to ensure high level of DTPR protein display on M13 phages.
VLR
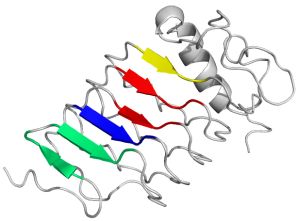
Variable Lymphocyte Receptors – unique antigen binding receptors in jawless vertebrates, are the oldest adaptive immune system molecules, which are not structurally similar to immunoglobulin-based antibodies. The VLRs consist of a leucine-rich repeat (LRR) which form a curved solenoid, and its concave surface is formed from β strands and has been suggested as the antigen-binding site. The convex (outer) surface is composed of the more diverse secondary structure elements.
Based on crystal structures of VLRs with its ligands, protein-protein interaction aminoacid positions at concave surface have been chosen to design phage display libraries.
PB1 – 16 positions, amino-acid size of 1×1012
ST – 11 positions, amino-acid size 1×108